How are Stanford's STARR database,the OMOP Common Data Model, and Epic's EHR Related?
Overview of Epic, OMOP CDM, and Stanford's STARR database, and how can to use them for EHR research
In this post, I explain what Epic (Chronicles, Clarity, etc.), OMOP CDM, and Stanford STARR are, how they are all related, and what the benefits/features of each are.
Hopefully this is helpful to any Stanford researcher or student dipping their toes into electronic health record (“EHR” or “EMR”) data for the first time, and aren’t clear how all these pieces connect.
In a nutshell:
- Epic is the EHR that records raw healthcare data.
- This is what your doctor interacts with every day.
- Stanford STARR is a SQL database purpose-built for researchers.
- Doctors do not use or see this.
- It is hosted in the cloud on Google Cloud BigQuery
- STARR extracts data from Epic and cleans it up nicely into a database with a simpler structure (i.e. “schema”)
- OMOP CDM is the name of the SQL schema specification that Stanford STARR follows.
- Think of it like a blueprint for organizing EHR data.
- Other universities have their own implementations of OMOP CDM as well.
- You may hear it be referred to interchangeably as “OMOP CDM” or just “OMOP”.
Most of this post is taken from the References at the bottom of this post – these are fantastic resources, so please check them out!
Epic
Epic is a popular vendor of electronic health records (EHRs).
As the EHR itself, the data that Epic collects is healthcare data in its rawest, purest, messiest, most real-time form. All of the other databases described in this post (OMOP CDM and STARR) are simplified databases that extract data from Epic and transform it into something more manageable for research/study purposes.
Here is a detailed overview of Epic’s architecture:
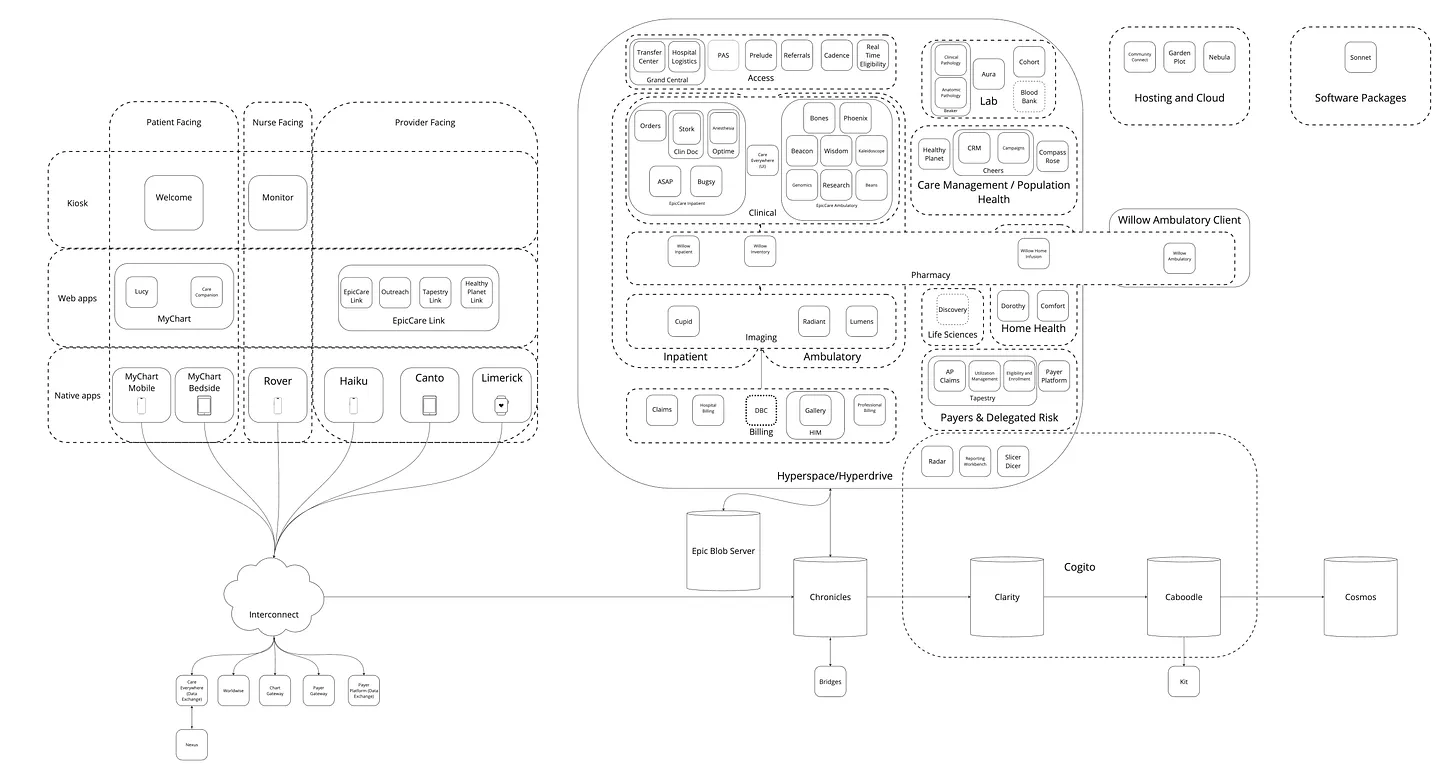
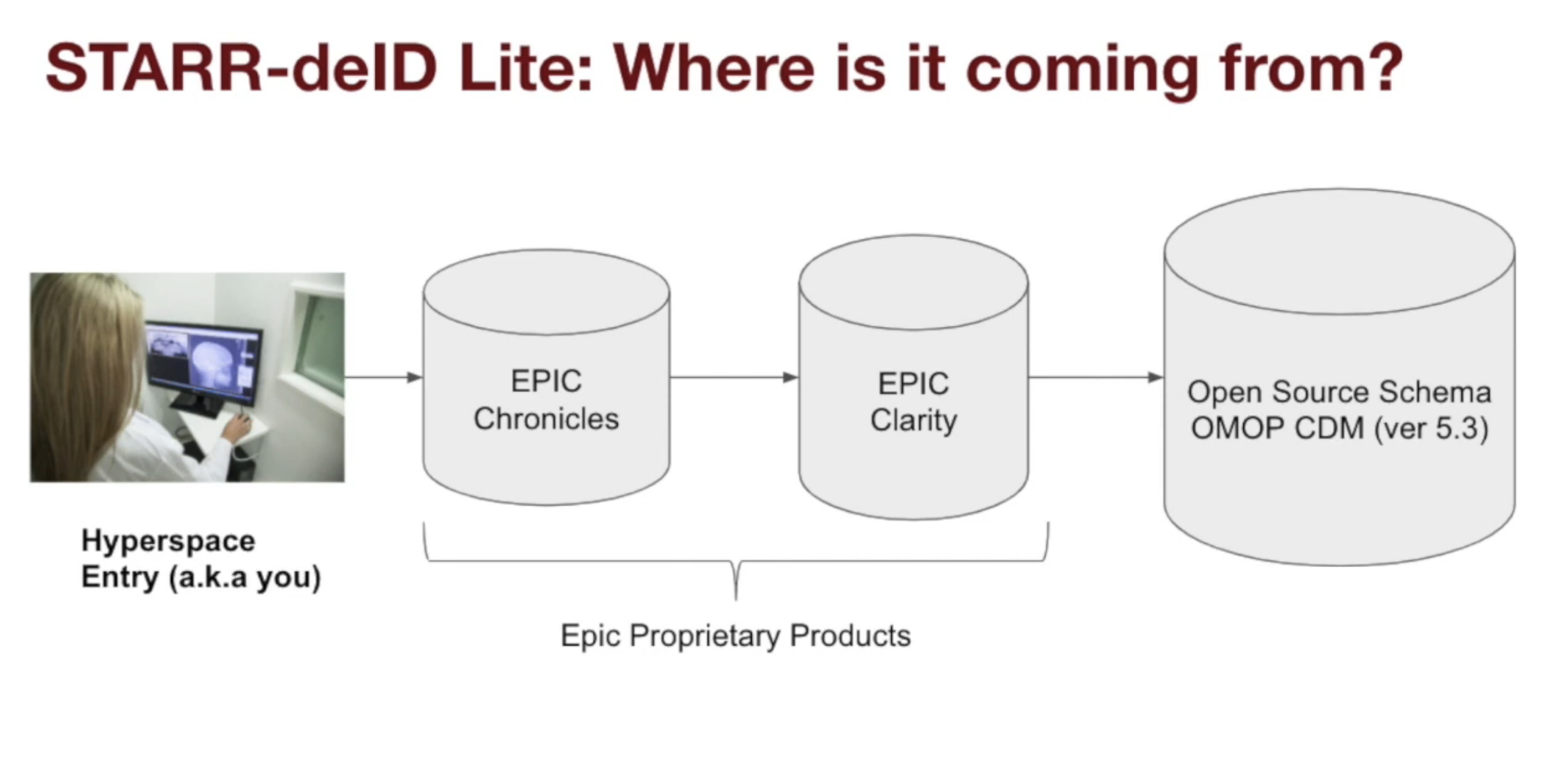
We’ll focus on their three most relevant products for research data analyses: Hyperspace (a GUI), Chronicles (a database), and Clarity (a database).
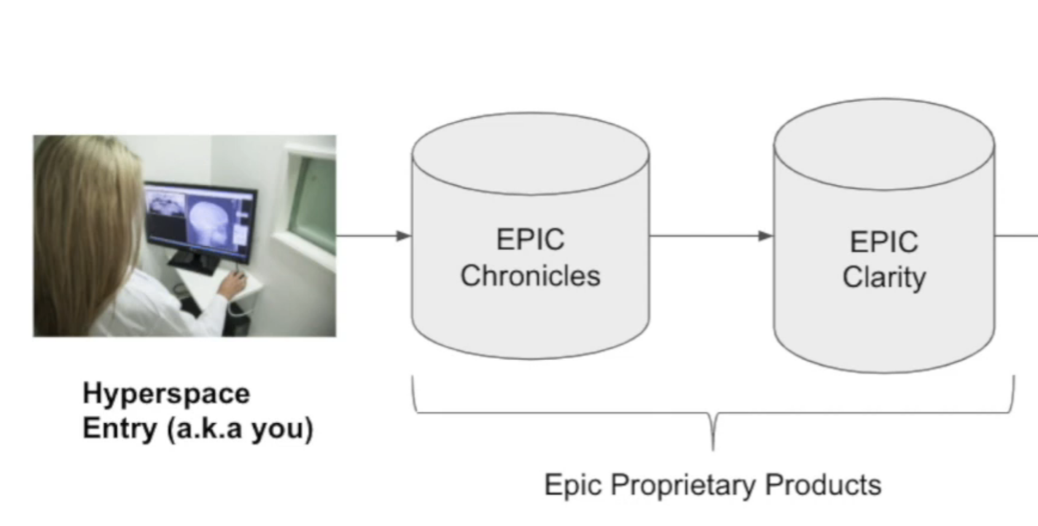
Hyperspace (GUI App)
- GUI application that doctors interact with
- Can pull in data from Epic Chronicles + other non-Epic databases (e.g. CT scans, pathology slides, etc.) and show to the doctor in one unified interface
- NOTE: Not all data visible in Hyperspace exists in Chronicles (and thus won’t be imported into Clarity/STARR)
Chronicles (Database)
- Hierarchical database (written in MUMPS)
- Designed for drilling into specific individual patients
- Not relational
-
Direct interface with day-to-day operations
- Core database of the EHR
Clarity (Database)
- Relational database (SQL)
- Losslessly extracts data from Chronicles
- Used for summary statistics
OMOP CDM
Definition
The OMOP (Observational Medical Outcomes Partnership) Common Data Model (CDM) is a standardized EHR database schema defined by the OHDSI (Observational Health Data Science & Informatics) consortium.
Why OMOP CDM?
Healthcare data is messy. The OMOP CDM solves this by creating a standard format for working with it.
Every EHR and every hospital has a unique schema for its databases, and the specific entities stored within those databases also differ across hospitals (i.e. some use ICD-10 codes for billing, others ICD-9 codes, etc.).
As a concrete example, the below image shows five different datasets, where each row represents the same type of event being recorded in each of the five datasets.
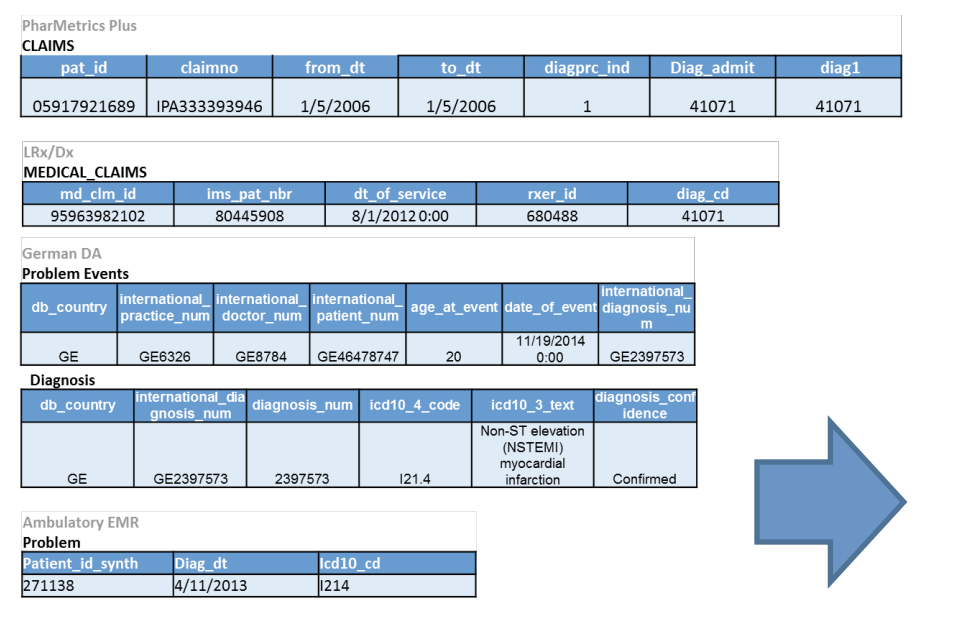
Note that each database has a different number of columns, column names, and cell values, even though they all encode the same event!
Now, let’s see what these tables look like once they’ve been converted into the OMOP CDM format:
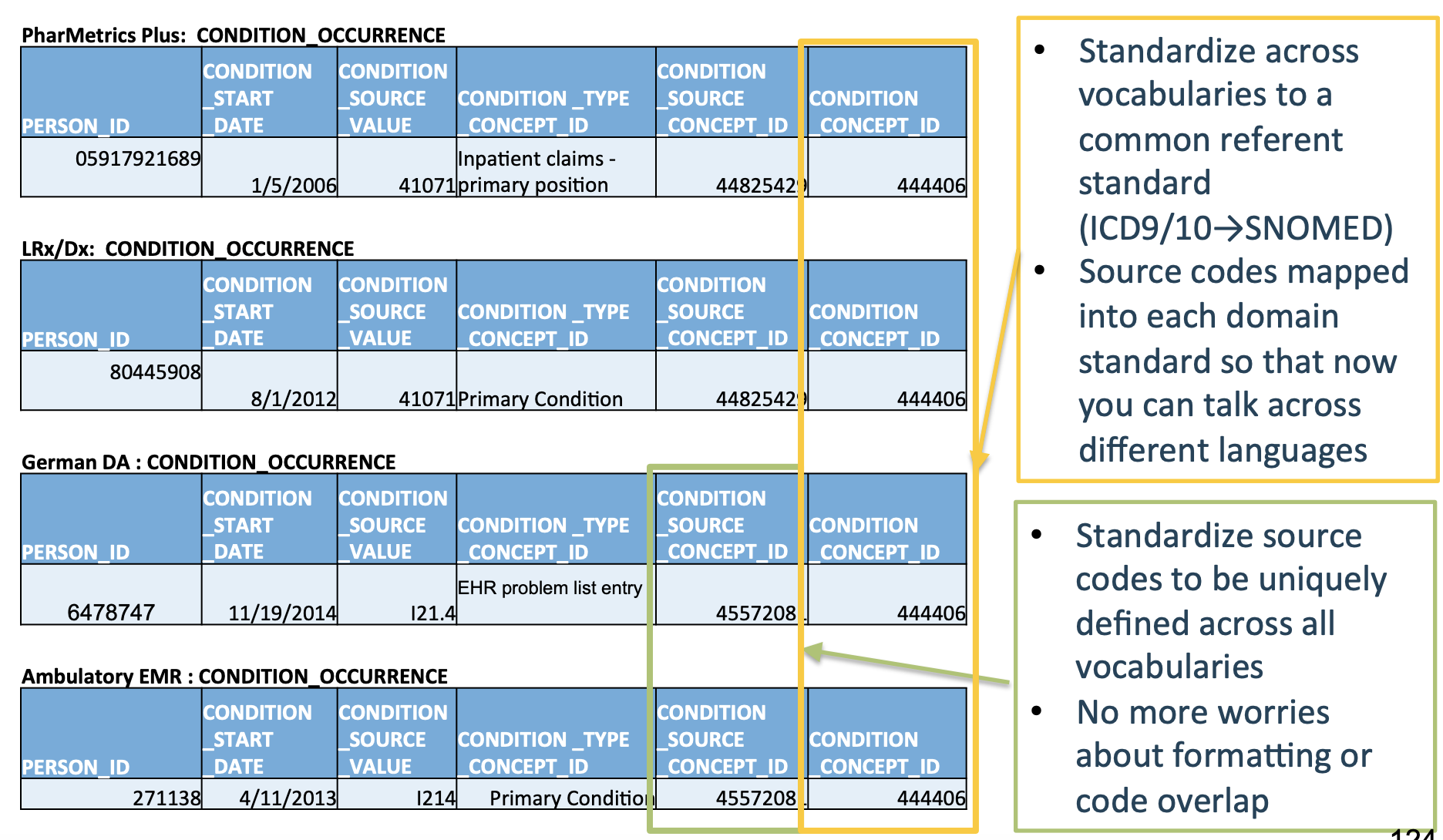
Much cleaner!
OMOP CDM creates a standard schema (“common data model”) and a standard set of medical terms (“common representation”) so that studies done at one hospital using the OMOP CDM can instantly be applied to another hospital using the OMOP CDM.
As the OHDSI community page outlines:
“The OMOP Common Data Model allows for the **systematic analysis of disparate observational databases [i.e. different hospitals’ EHRs] **.
The concept behind this approach is to transform data contained within those databases into a common format (data model) as well as a common representation (terminologies, vocabularies, coding schemes), and then perform systematic analyses using a library of standard analytic routines that have been written based on the common format.
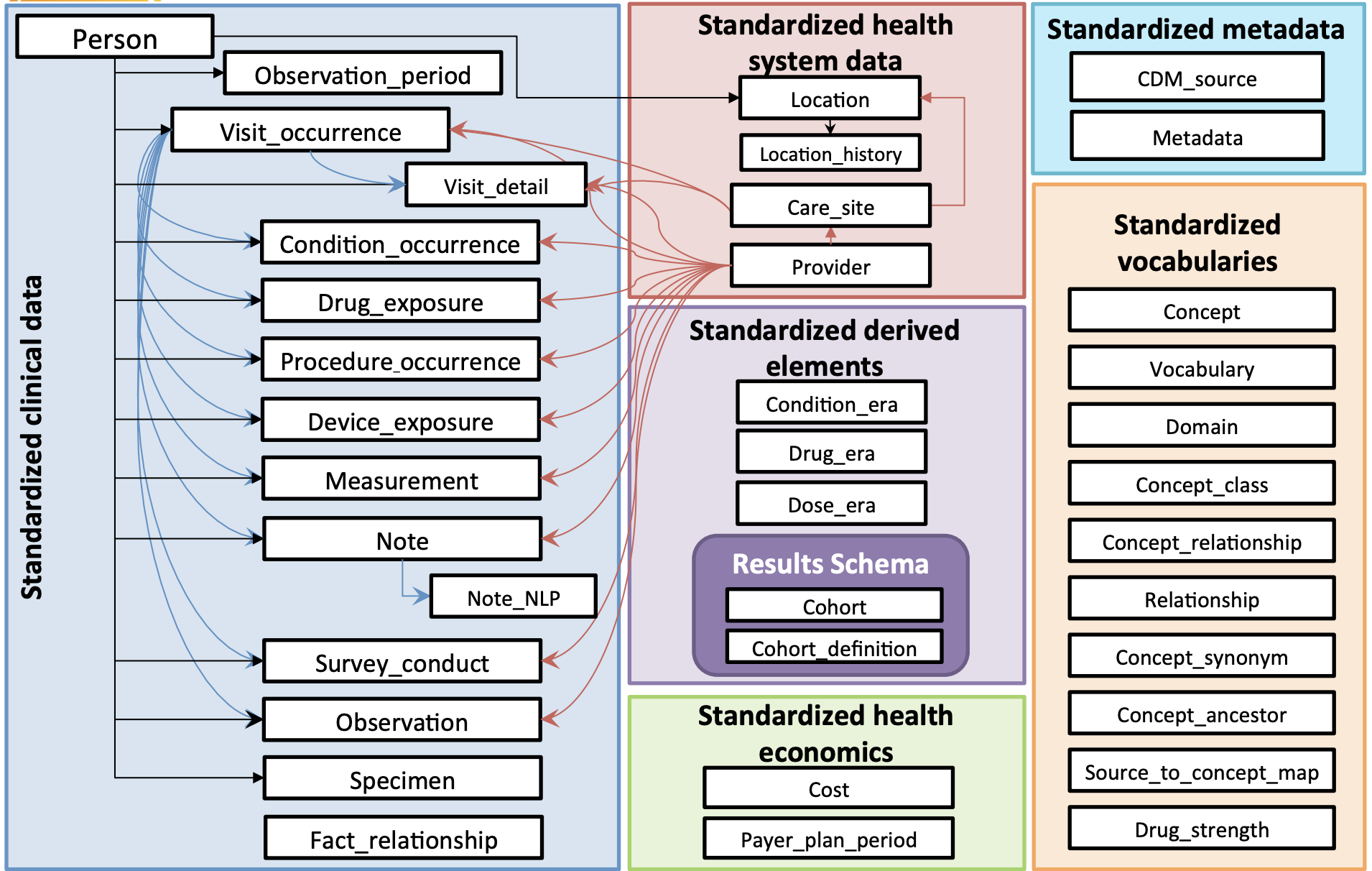
OMOP Schema
OMOP CDM is a SQL schema comprised of…
- 37 tables
- 394 fields within those tables
The below chart shows all of the tables and their relationships. Each white box is a table:
OMOP Terms
Concepts
A “concept” is any sort of entity in your data (diseases, diagnoses, procedures, medical terms, etc.).
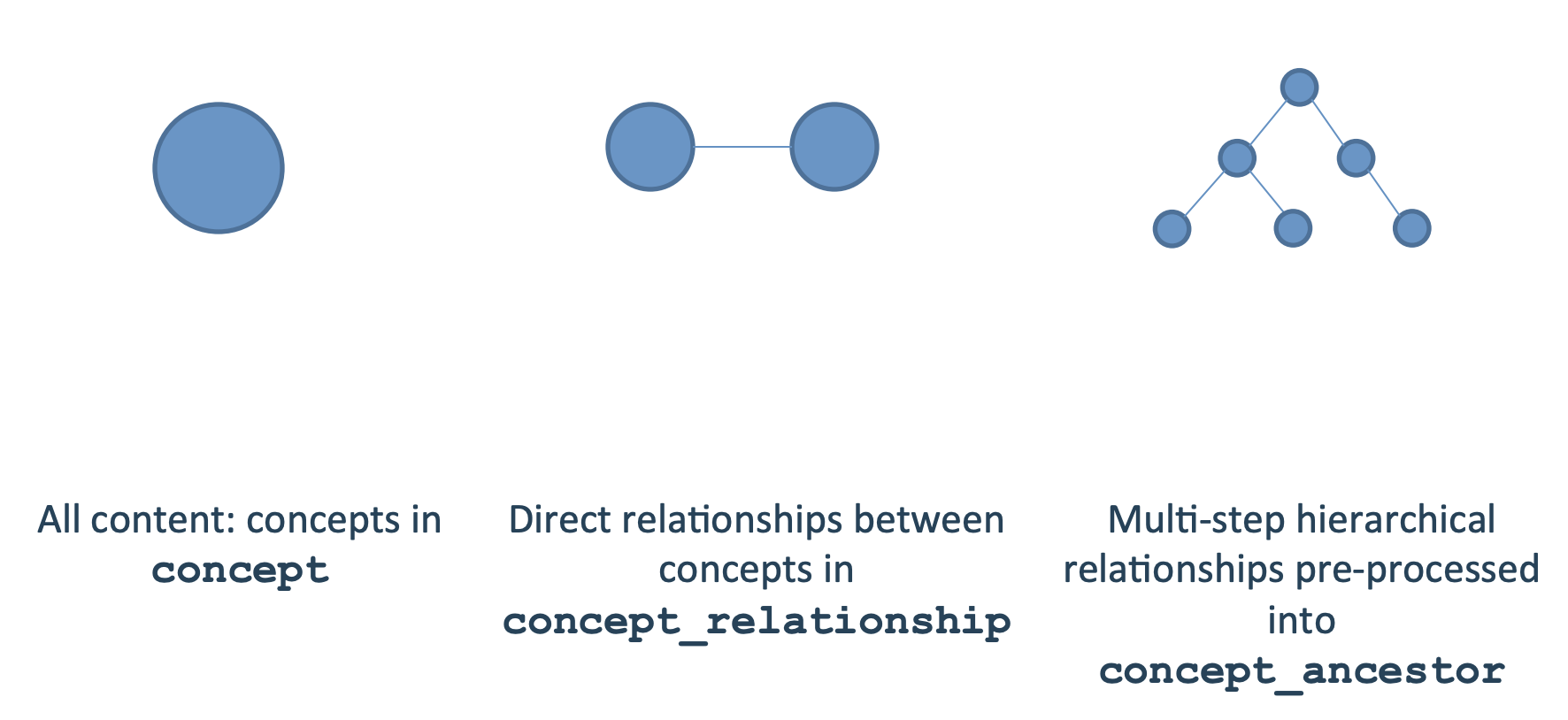
Information on concepts is stored in three tables in the OMOP CDM:
- concept - stores all concepts
- concept_relationship - stores direct relationships between concepts
- concept_ancestor - precomputes in-direct relationships between concepts
Here is what the concept table looks like for Stanford’s OMOP CDM instance (“STARR”):
And here is an example of a specific concept – Atrial Fibrillation (313217)

concept_id: Unique OMOP ID for this concept (313217)concept_name: Human readable English description (Atrial fibrilation)domain_id: Domain to which this concept belongs (Condition)standard_concept: If “S”, then this is the standard concept for this entity in OMOP (S)vocabulary_id: Vocabulary this concept came from, e.g. SNOMED, ICD, LOINC, etc. (SNOMED)- Comes from original vocabulary
concept_class_id: Class of this concept in its original (i.e. source) vocabulary (Clinical Finding)- Comes from original vocabulary
concept_code: Code for this concept in its original (i.e. source) vocabulary (49436004)- Comes from original vocabulary
- Not unique across concepts (e.g. there are 9 unique OMOP concepts with a
concept_code = 1001)
valid_start_date, valid_end_date: Concept is valid during this time interval (2002-01-31 to 2099-12-31)- Useful because concepts can enter / leave the vocabulary over time, e.g. in 2019 the concept COVID-19 entered the vocabulary
Note: Athena is a useful web GUI provided by OHDSI for searching through all OMOP concepts.
Domain
A “domain” is the OMOP “type” of a concept. Examples include condition, drug, device, measurement, procedures, diagnoses.
Vocabulary
A vocabulary is an ontology of concepts. Examples include SNOMED, ICD, MESH, RxNorm, CPT, etc.
A standardized vocabulary is the 1+ vocabular(ies) for each domain that take(s) primacy over all other vocabularies. All of the concepts within a domain will be mapped to a concept from that domain’s standardized vocabulary.
- NOTE: The standardized vocabulary can be composed of one or more vocabularies for a domain, when one vocabulary alone is insufficient to provide sufficient coverage. See
proceduresbelow.
Examples:
- For
diagnoses, SNOMED is the standard vocab. For example, all ICD/SNOMED/MESH codes for atrial fibrilation get mapped to the SNOMED code for atrial fibrilation. - For
procedures, there are multiple vocabularies that are part of the standardized vocabulary (i.e. CPT + SNOMED + others) - For
drugs, RxNorm is the standard vocab
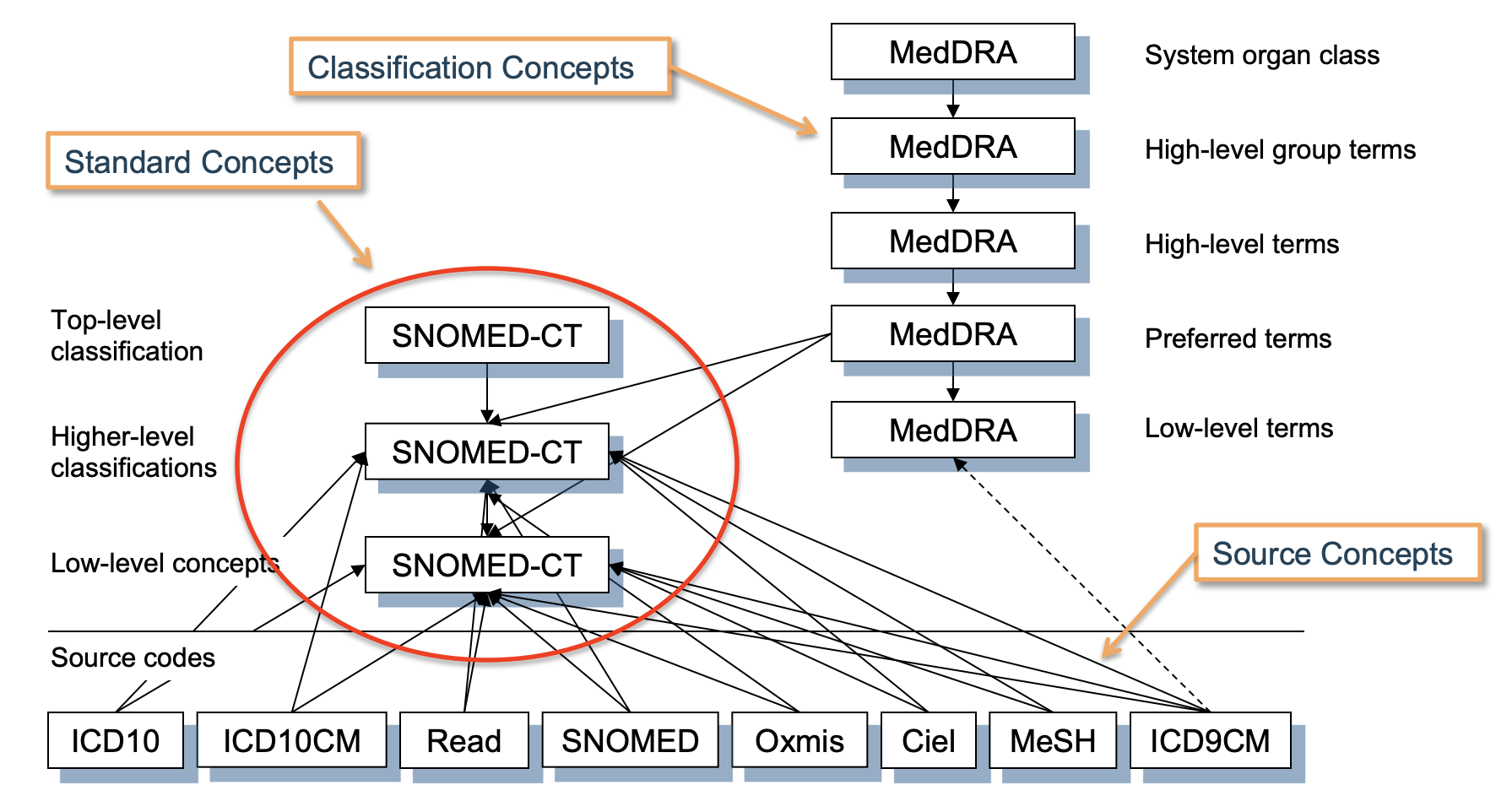
How to map Source Concepts -> OMOP Standard Concepts
Here, we’ll walk through an example taken from the OHDSI training materials.
Goal: We’re given the ICD-9 concept 427.31 (atrial fibrilation). How do we get its corresponding standard OMOP concept?
Steps:
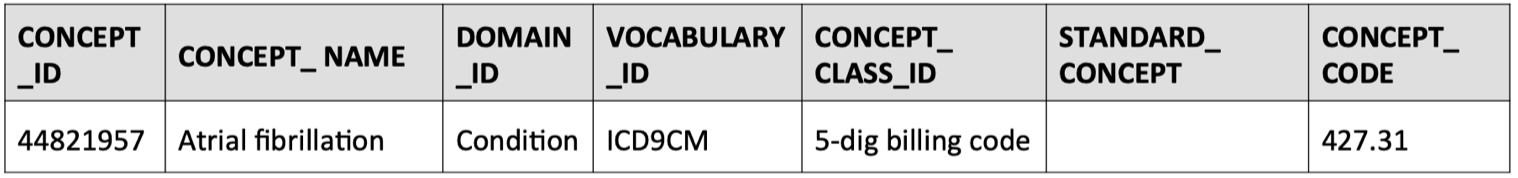
- Find the source concept ID in the concept table that corresponds to the
concept_code = 427.31, which in this case is given by:concept_id = 44821957
SELECT * FROM concept WHERE concept_code = '427.31'
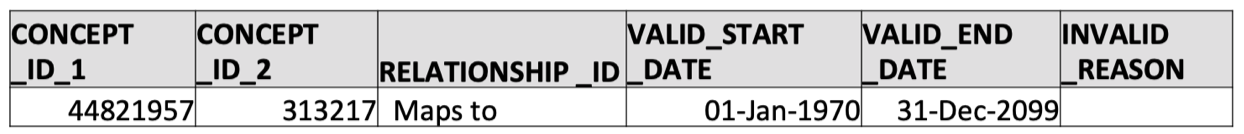
- Map the source concept ID to its corresponding standard concept ID using the concept_relationship table. Note that the
relationship_id = 'Maps to'maps a non-standard concept (labeled asconcept_id_1) to its OMOP standard concept (labeled asconcept_id_2)
SELECT * FROM concept_relationship WHERE concept_id_1 = 44821957 AND relationship_id = 'Maps to';
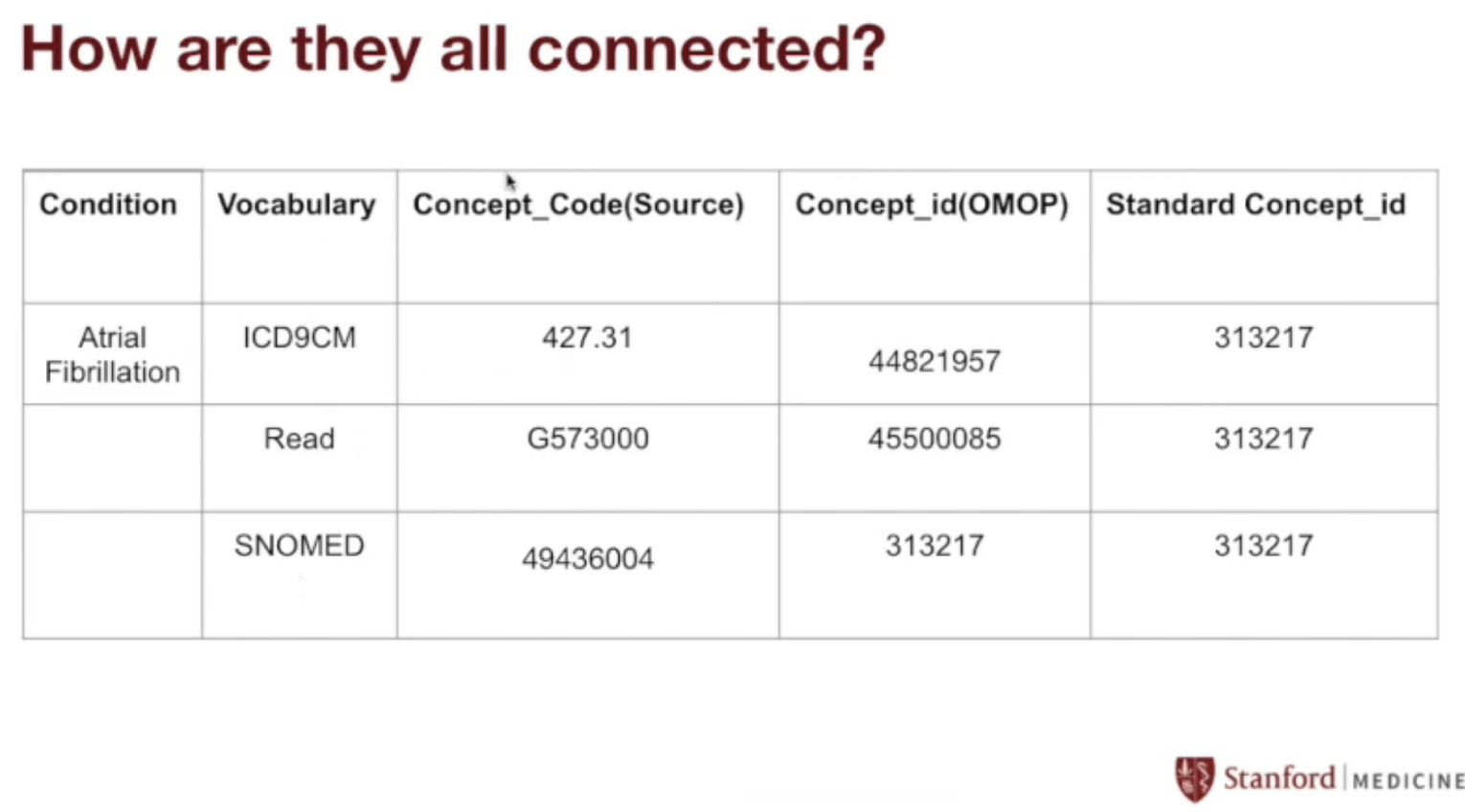
- That’s it! We now know that the OMOP standard concept ID corresponding to ICD-9 code 427.31 (atrial fibrilation) is 313217.
Generalizing:
This process works for every vocabulary.
We started with the ICD-9 code for atrial fibrilation (427.31), but we could also have used the Read code for atrial fibrilation (G573000), or the SNOMED code for atrial fibrilation (49436004).
The amazing thing about the OMOP CDM is that they all map to the same standard concept ID of 313217, which makes sense since they all refer to the same thing!
NOTE: Concepts in other OMOP CDM tables will contain a reference to both the standardized concept_id as well as the original non-standard source concept as source_concept_id to preserve data provenance.
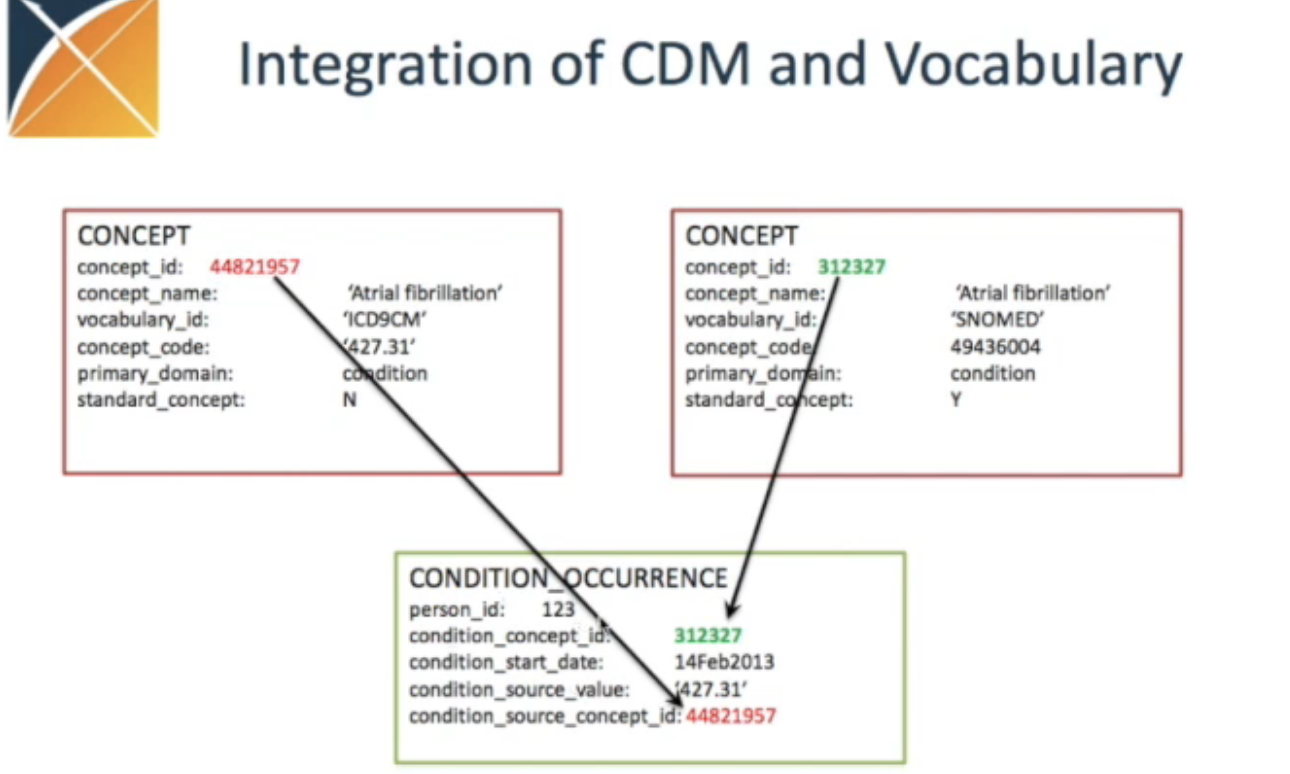
Types of concept relationships
-
“A Subsumes B” => A is a parent of B
-
“A Is-A B” => A is a child of B
-
“Levels of separation” between A and B => Number of Is-A or Subsumes relationships between A and B
Stanford STARR
Stanford’s version of OMOP CDM is called STARR-OMOP (STAnford Research Repository).
Stanford uses v5.3 of the OMOP CDM specification.
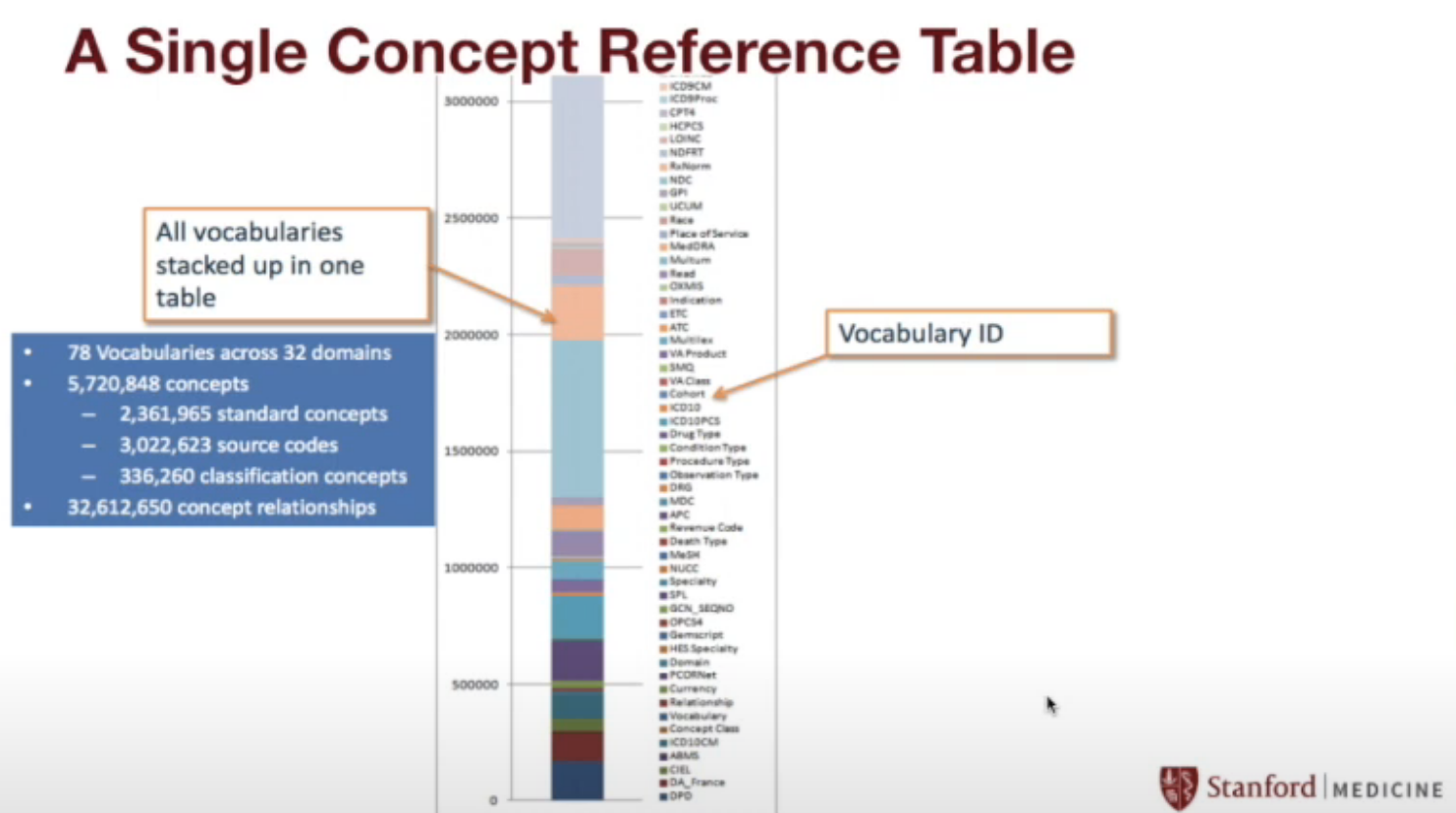
STARR by the numbers
STARR contains…
- OMOP Concepts…
- 78 vocabularies
- 32 domains
- 5.7M concepts
- 32M concept relationships
- Raw Data…
- 345GB of clinical notes
- 156M unique notes
- 33B words
- 3.5M unique patients
- 14TB of pediatric vitals
- 345GB of clinical notes
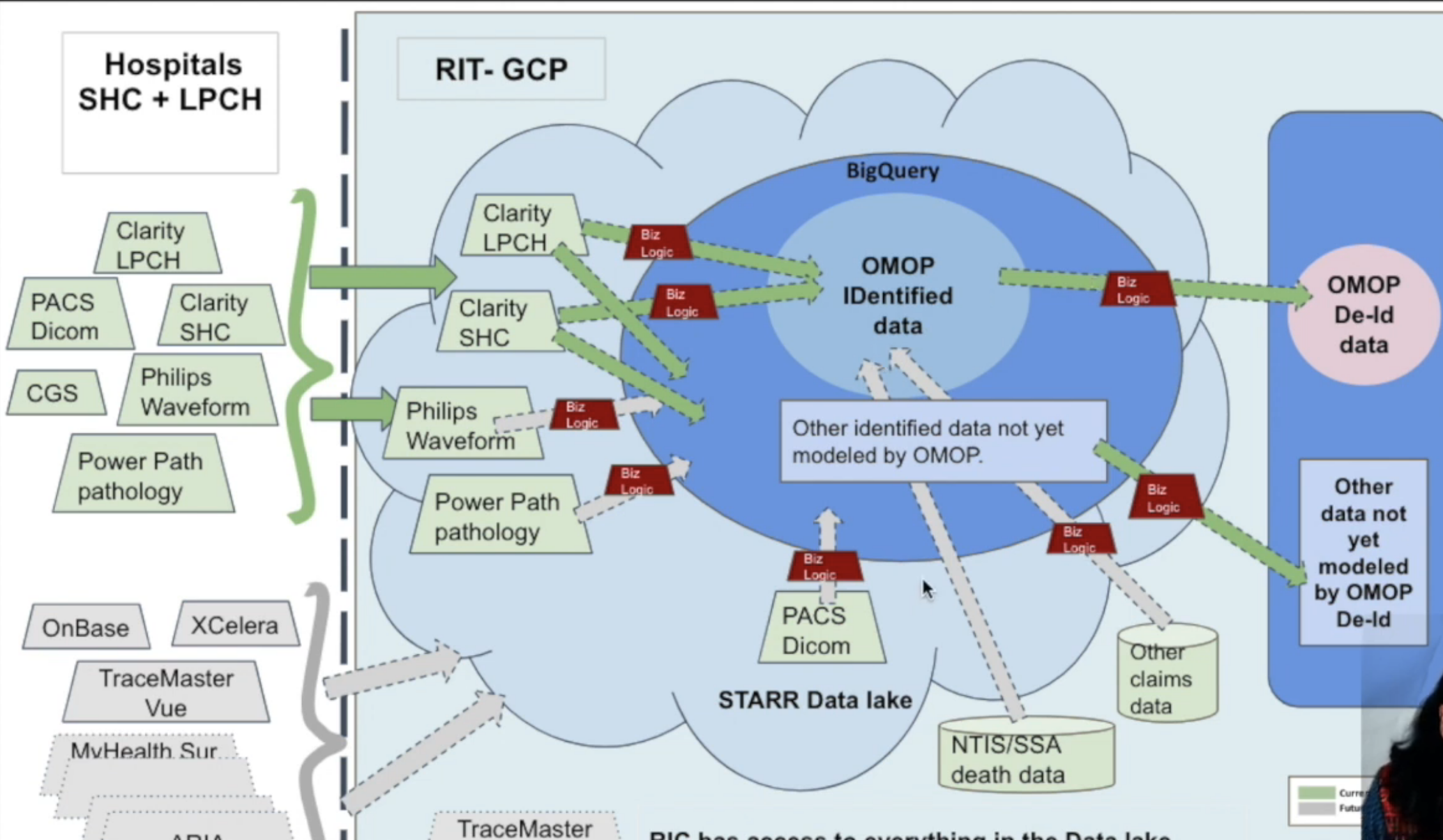
Data Generation Process
Stanford has build its own internal ETL pipeline that maps data from Epic Clarity into STARR-OMOP.
There are multiple versions of STARR-OMOP, as listed below. Red = high risk, Yellow = medium risk data
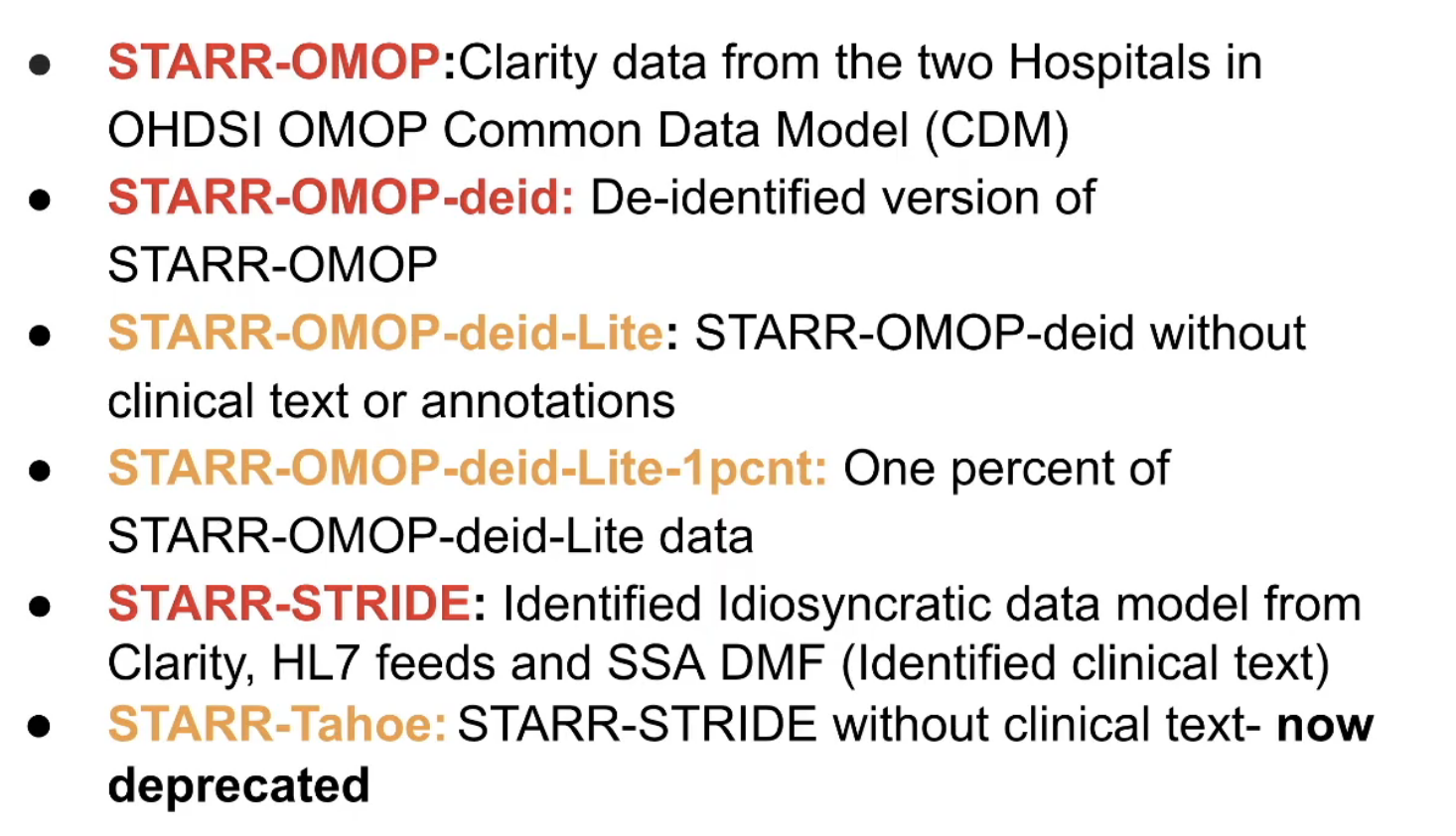
Some notes:
- STARR-OMOP is stored in Google Cloud BigQuery
- STARR-OMOP-deid is available without IRB approval to Stanford researchers
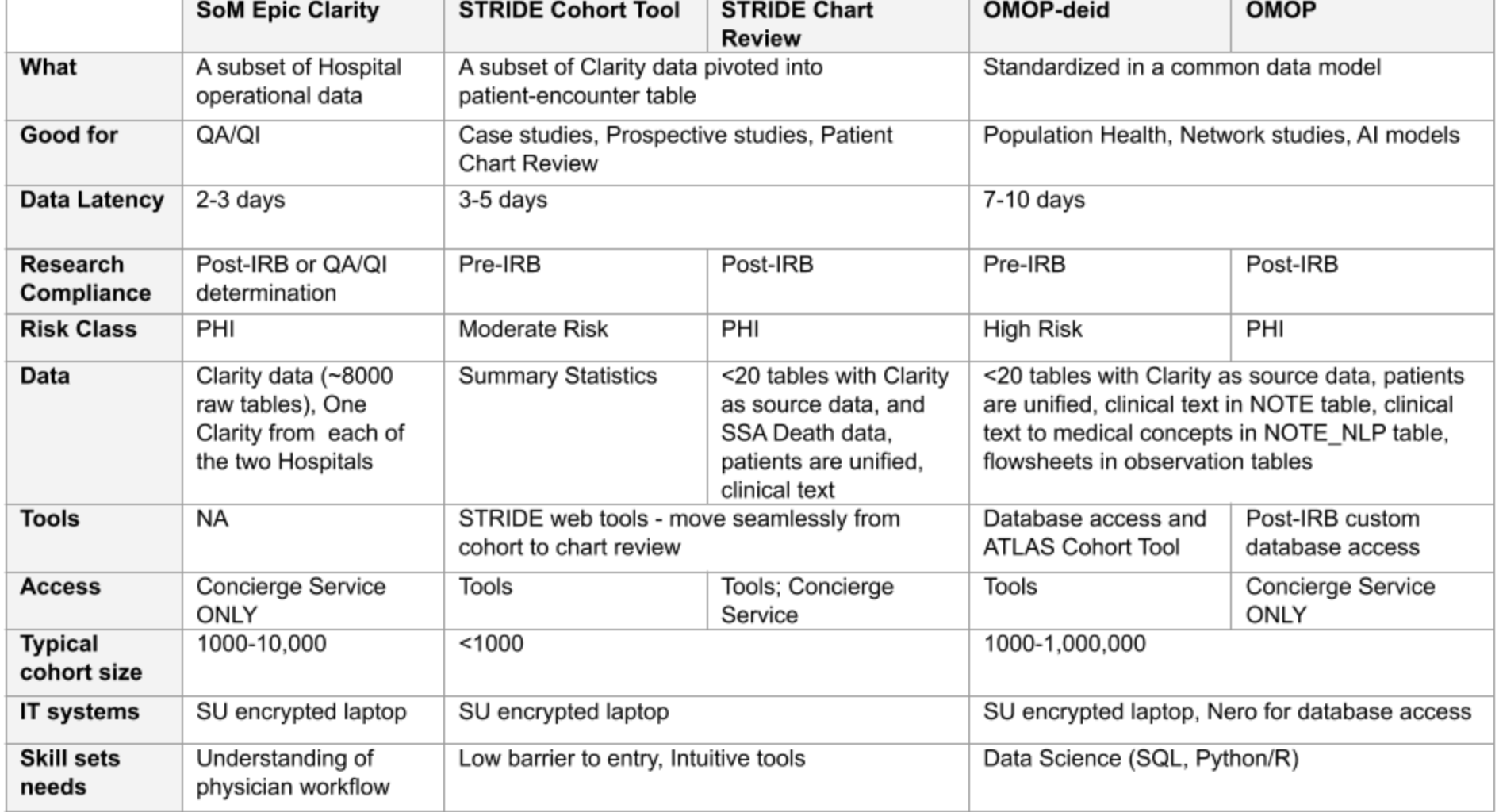
Differences between Clarity, STRIDE, and STARR-OMOP
STRIDE is the old Stanford data warehouse (which STARR replaced).
A helpful table from the Stanford Research IT website outlines their main differences:
NOTE_TEXT Table
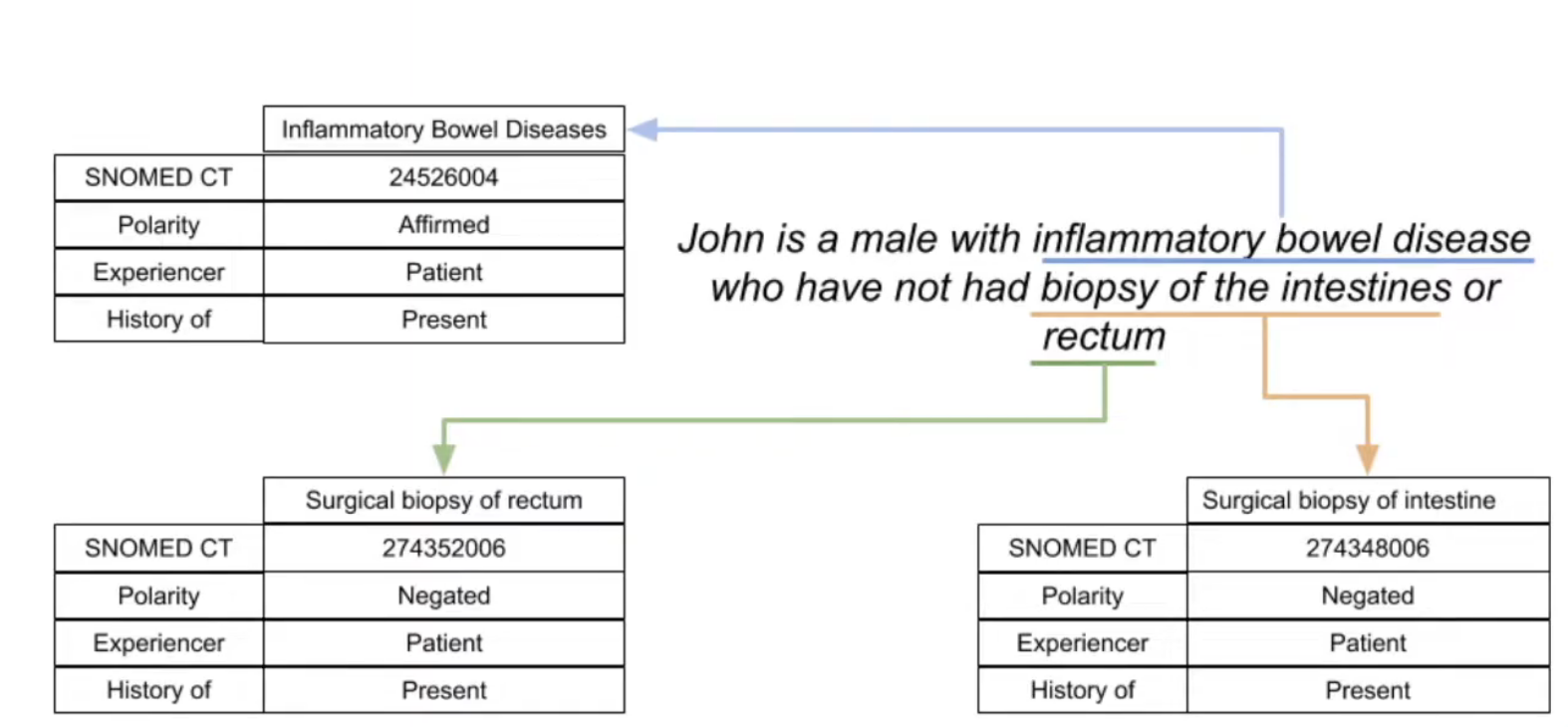
A unique aspect of STARR-OMOP is the NOTE_TEXT table, which contains a bunch of natural language processing (NLP) features that are automatically extracted from the textual content of the NOTE table (which contains all clinical notes written at Stanford Hospital).
Some notes about this table:
- It uses the ConTex + Negex pipelines to generate features
- A single note can contain multiple annotations
- Only supports exact string matches