Walkthrough of the OMOP CDM (Part 1)
Defining core concepts in the OMOP CDM
In Part 1 of this series on the OMOP CDM, we explain what the OMOP CDM is, define key terms such as “concept”, “source value”, “vocabulary”, and “domain”, and describe how they related to each other.
This post is organized as follow:
- Motivation
- Technical Overview of OMOP CDM
- Terminology + Key Concepts
- Domains, Vocabularies, and Concepts
- Field Naming Conventions
- Read / Write Permissions
Motivation
Problem: Healthcare Data Lacks Standardization
Healthcare data is messy, and electronic health records (EHR) are notoriously incompatible.
Making health records “interoperable” has been a multi-decade battle.
Almost every EHR has a unique database set-up, and the specific values stored within those databases often differ across hospitals (i.e. some use ICD-10 codes for billing, others ICD-9 codes, etc.).
As a concrete example, the below image shows five different EHR datasets, where each row represents the same exact type of event being recorded:
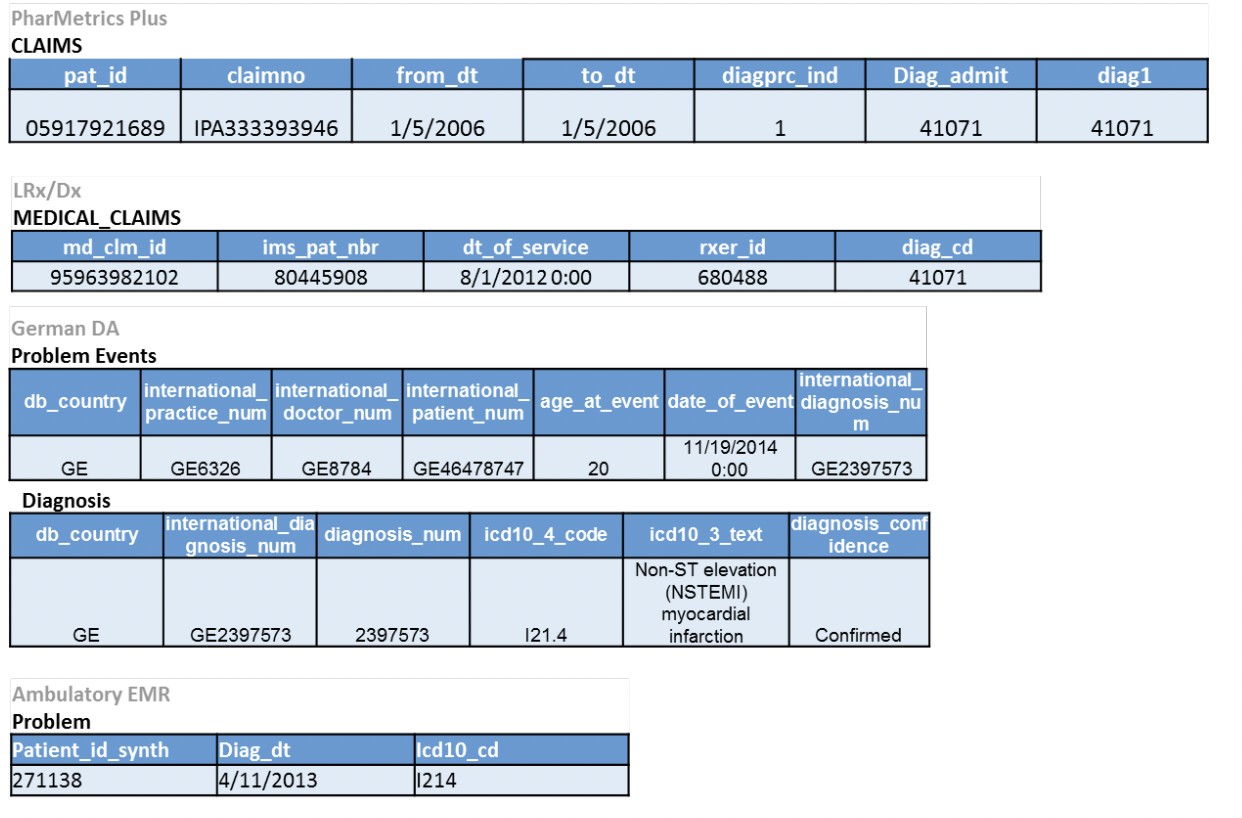
Note that each database has a different number of columns, column names, and cell values, even though they all encode the same event!
Solution: A “Common Data Model” (CDM)
The OMOP CDM solves this problem by creating a standard format for working with healthcare data.
More specifically, OMOP (Observational Medical Outcomes Partnership) Common Data Model (CDM) is a standardized EHR database schema defined by the OHDSI (Observational Health Data Science & Informatics) consortium.
Let’s see what the five aforementioned tables look like once they’ve been converted into the OMOP CDM format:
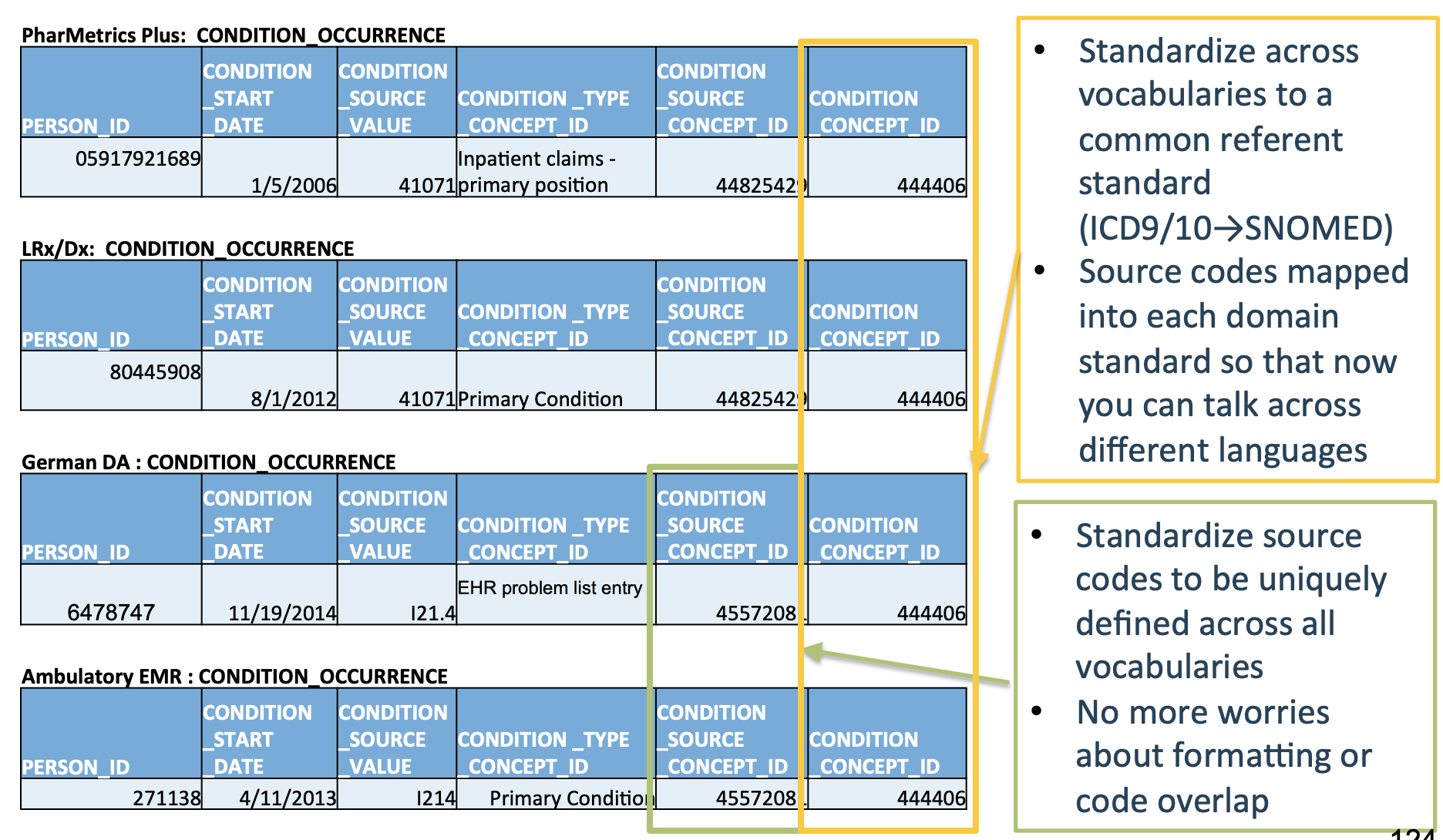
Much cleaner!
This enables research done at one hospital to easily transfer to any other hospital that also uses the OMOP CDM.
OMOP CDM creates a standard schema (“common data model”) and a standard set of medical terms (“common representation”) to improve healthcare data interoperability.
As the OHDSI community page outlines:
“The OMOP Common Data Model allows for the **systematic analysis of disparate observational databases [i.e. different hospitals’ EHRs] **.
The concept behind this approach is to transform data contained within those databases into a common format (data model) as well as a common representation (terminologies, vocabularies, coding schemes), and then perform systematic analyses using a library of standard analytic routines that have been written based on the common format.
Technical Overview of OMOP CDM
The OMOP CDM is a person-centric, relational database schema that contains 39 tables split over 7 main table categories.
The below image (taken from here) shows the structure of the OMOP CDM.
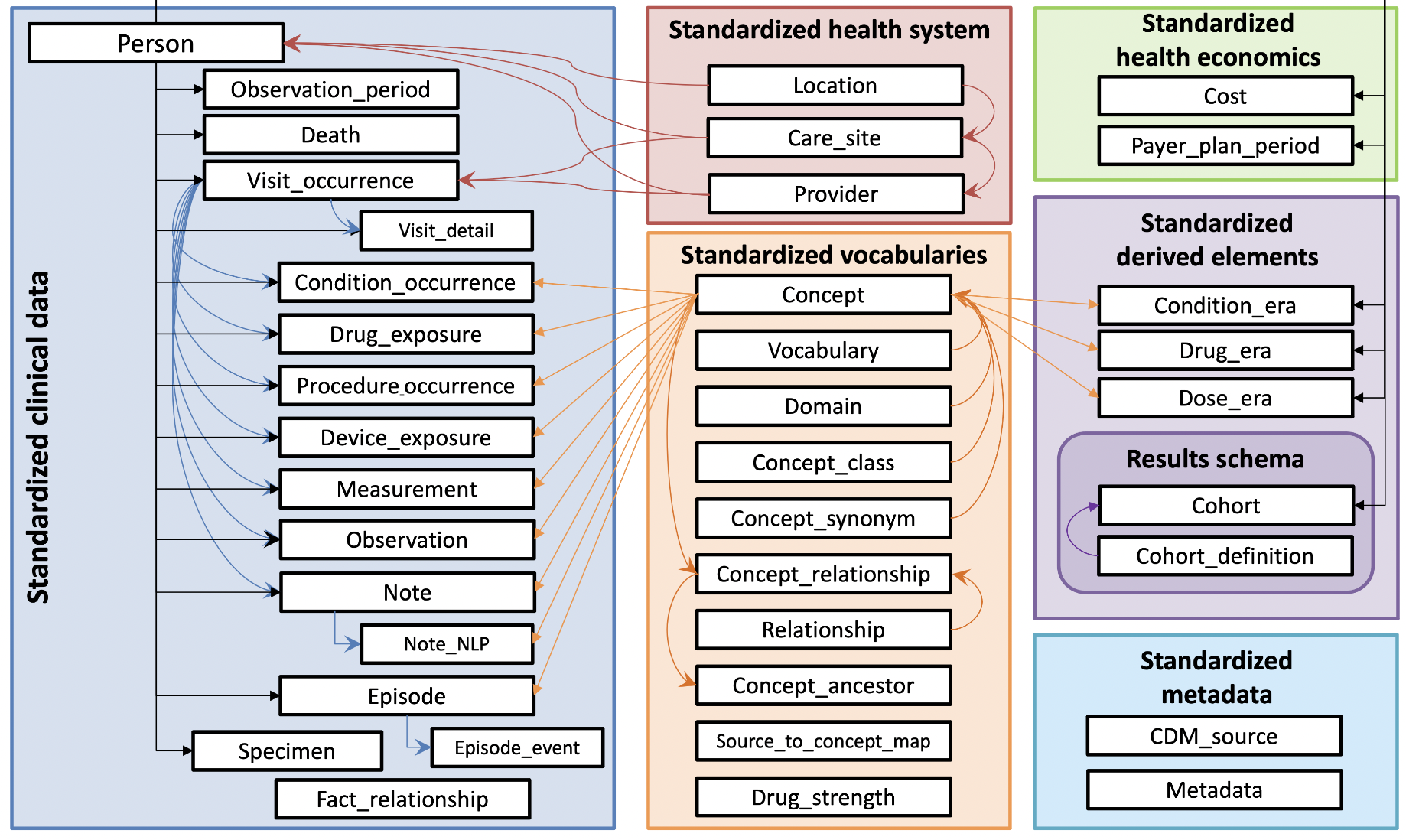
Let’s breakdown the bolded terms in the above sentence piece-by-piece.
39 tables
Each white box in the above image is a unique table. Each table contains a set of fields (i.e. columns) which have a specifically defined datatype, semantic meaning, and relationship to the other tables. The columns for all 39 tables are defined here.
7 main table categories
The tables are categorized into the following main groups:
| Table Name | Color | # of Tables |
|---|---|---|
| Clinical data | Blue | 16 |
| Vocabularies | Orange | 10 |
| Health system | Red | 4 |
| Derived elements | Light purple | 3 |
| Health economics | Green | 2 |
| Metadata | Turquoise | 2 |
| Results | Dark purple | 2 |
The Results tables are the only ones that are writable by end-users.
Person-centric
We take a person-centric view of healthcare, where the individual patient is our primary unit of analysis.
Every table containing clinical events (i.e. everything in the blue Clinical data square above) must be linked to a specific person in the PERSON table.
Relational Database Schema
The OMOP CDM is simply a blueprint (i.e. “schema”) for how data should be structured.
OMOP CDM does not require any particular software implementation – any type of relational database will suffice (e.g. MySQL, Postgres, SQL Server, Oracle, etc.)
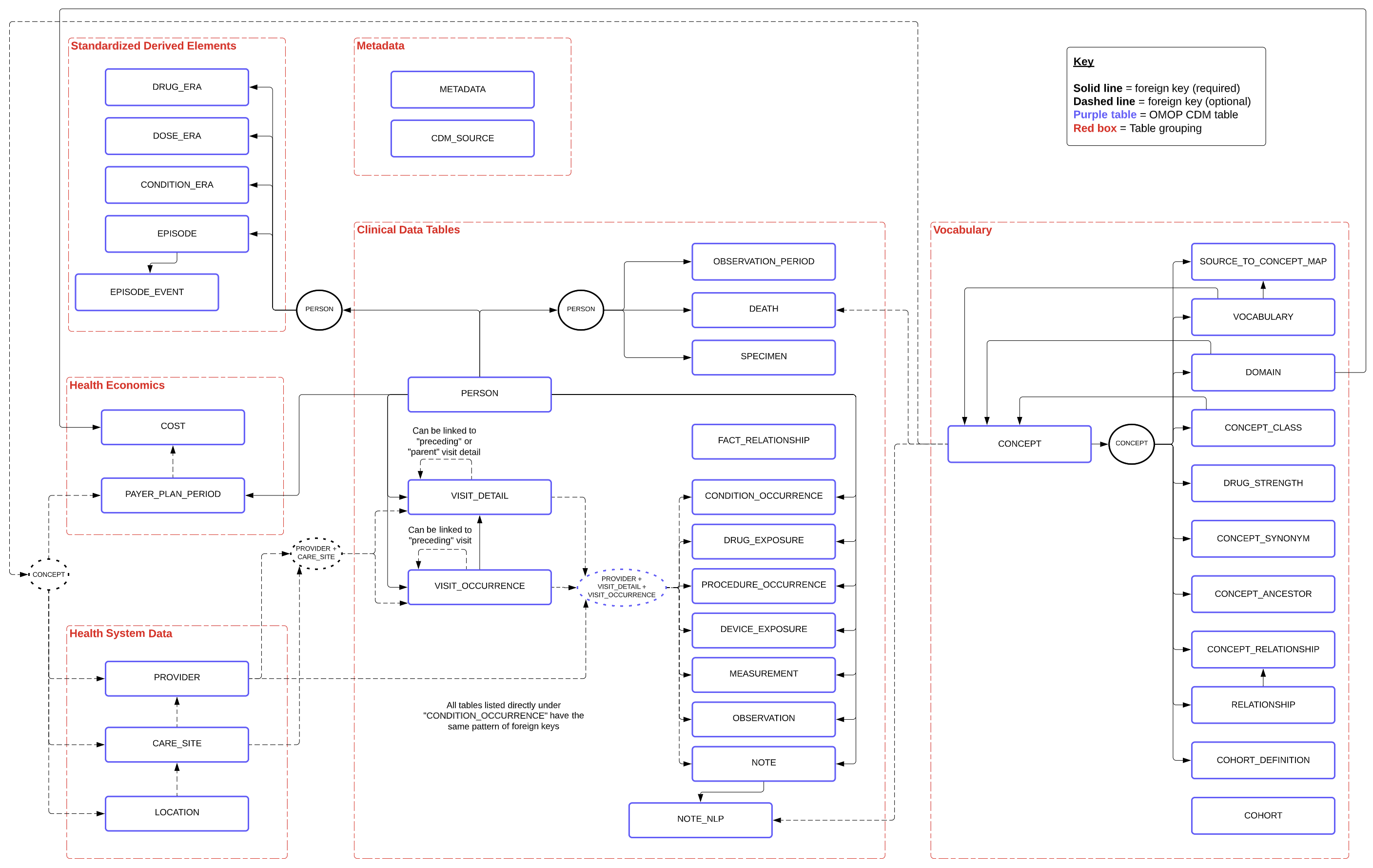
A more detailed view of the OMOP schema, with all foreign keys (required in solid black lines, optional in dashed lines) between tables, is shown below (using the alternative grouping described here):
Terminology + Key Concepts
tl;dr
A “concept” is any sort of entity in your data (diseases, diagnoses, procedures, medical terms, etc.).
The relationship between source values, concepts, domains, and vocabularies is as follows:
- A source value is the raw clinical event extracted from the EHR (“0.011”)
- The source value gets transformed into a source concept (“44828631” for ICD9CM/011 for pulmonary tuberculosis), which has associated with it one domain (e.g. “Condition”) and one vocabulary (e.g. “ICD9CM”)
- If a source value does not have a corresponding code in an existing vocabulary, it may be defined de-novo as a custom “OMOP” code, and would then just be referred to as a concept (since there’s no “source” vocab for that concept)
- If this is a non-standard concept, it then gets transformed into a standard concept (253954 for pulmonary tuberculosis, taken from SNOMED/154283005).
Here, we define each of these terms and *underline* their relationship to each other.
Source Value
A source value is the raw form that a clinical fact was represented in the original source EHR.
- Usage
- The
[EVENT]_SOURCE_VALUEcolumn of tables
- The
- Example: 011 – This could be ICD9CM code for “Pulmonary Tuberculosis”, the DRG code for “Nervous System Neoplasms without Complications, Comorbidities”, or the UB04 code for “Hospital Inpatient (Including Medicare Part A)”
A source value gets transformed into a...**source concept**
Source Concept
A source concept is an OMOP-specific entity that “normalize[s] the meaning of a clinical fact.” In other words, it is a fully standardized, uniform representation of a clinical fact from the source (Note: This occurs before conversion to a standard concept, as described below)
- Definition
- Table: CONCEPT
- Fields (shown in table below)
- Usage
- In the
[EVENT]_SOURCE_CONCEPT_IDcolumn of tables as foreign keys to the CONCEPT table
- In the
- Example
- 44828631 – This is unambigously ICD9CM/011 for “Pulmonary Tuberculosis”, and belongs to the domain “Condition”
| Field | Example Value | Note |
|---|---|---|
| CONCEPT_ID | 313217 | Primary key (globally unique) |
| CONCEPT_NAME | Atrial fibrillation | English description |
| DOMAIN_ID | Condition | Single alphanumeric string for its one domain |
| VOCABULARY_ID | SNOMED | Vocab this concept came from |
| CONCEPT_CLASS_ID | Clinical finding | Class in original vocab (SNOMED) |
| STANDARD_CONCEPT | S | If S , then standard concept. Otherwise, NULL |
| CONCEPT_CODE | 49436004 | Code in original vocab (SNOMED) |
| VALID_START_DATE | 01-Jan-1970 | Time interval code is valid |
| VALID_END_DATE | 31-Dec-2099 | Time interval code is valid |
| INVALID_REASON | Time interval code is valid |
A source concept gets transformed into a...**standard concept** (sometimes).
Standard Concept
A standard concept is the single concept chosen for usage across the OMOP CDM to represent a clinical fact, when multiple source concepts from different vocabularies are synonymous. This ensures normalization across analyses.
- Definition
- Table: CONCEPT
- Fields:
- Have a
Svalue in the columnSTANDARD_CONCEPT
- Have a
- Usage
- In the
[EVENT]_CONCEPT_IDcolumn of tables as foreign keys to the CONCEPT table
- In the
- Example
- 253954 – This is unambigously the standard concept for “Pulmonary Tuberculosis.” It comes from the concept SNOMED/154283005, and is used instead of 44828631 (which is the ICD9CM/011 code for pulmonary tuberculosis)
Each concept belongs to a single...**domain**
Domain
A domain is a collection of concepts. It is a generic classification of the type of a concept (e.g. “Drug”, “Device”, “Measurement”).
- Domains appear in the
DOMAIN_IDcolumn of the CONCEPT table- Each domain is a short case-sensitive alphanumeric string
- One-to-many relationship from a domain -> concepts
Each concept also belongs to a single...**vocabulary**
Vocabulary
A vocabulary is also a collection of concepts. It represents the source where the corresponding concept was defined (e.g. “ICD9CM”). Thus, concepts in the same vocabulary can span many different domains.
- Vocabularies appear in the
VOCABULARY_IDcolumn of the CONCEPT table- Each vocabulary is a short case-sensitive alphanumeric string
- One-to-many relationship from a vocabulary -> concepts
Domains, Vocabularies, and Concepts
I’ve include more detail below on domains, vocabularies, and concepts for the curious reader.
Domains
There are a total of 30 domains in OMOP CDM.
They are defined in the DOMAIN table.
DOMAIN_ID= primary key – unique alphanumeric string of <=20 chars (e.g. “Drug”, “Device”)- Linked to concepts as a foreign key on the
DOMAIN_IDfield in the CONCEPT table
A list of all domains and their associated concepts is reproduced below (taken from here):
| Domain | Concept Count | Domain (cont.) | Concept Count (cont.) |
|---|---|---|---|
| Drug | 1731378 | Route | 183 |
| Device | 477597 | Currency | 180 |
| Procedure | 257000 | Payer | 158 |
| Condition | 163807 | Visit | 123 |
| Observation | 145898 | Cost | 51 |
| Measurement | 89645 | Race | 50 |
| Spec Anatomic Site | 33759 | Plan Stop Reason | 13 |
| Meas Value | 17302 | Plan | 11 |
| Specimen | 1799 | Episode | 6 |
| Provider Specialty | 1215 | Sponsor | 6 |
| Unit | 1046 | Meas Value Operator | 5 |
| Metadata | 944 | Spec Disease Status | 3 |
| Revenue Code | 538 | Gender | 2 |
| Type Concept | 336 | Ethnicity | 2 |
| Relationship | 194 | Observation Type | 1 |
Vocabularies
There are a total of 111 vocabularies currently supported by OHDSI, of which 78 are externally developed and 33 were internally developed by OHDSI.
They are defined in the VOCABULARY table.
VOCABULARY_ID= primary key – unique alphanumeric string of <=20 chars (e.g. “SNOMED”, “ICD9CM”)- Linked to concepts as a foreign key on the
VOCABULARY_IDfield in the CONCEPT table
A standardized vocabulary is the 1+ vocabular(ies) for each domain that take(s) primacy over all other vocabularies. All of the concepts within a domain will be mapped to a concept from that domain’s standardized vocabulary.
The table below shows the standardized vocabularies for some domains, taken from here:
| Domain | Standard Concepts |
|---|---|
| Condition | SNOMED, ICDO3 |
| Procedure | SNOMED, CPT4, HCPCS, ICD10PCS, ICD9Proc, OPCS4 |
| Measurement | SNOMED, LOINC |
| Drug | RxNorm, RxNorm Extension, CVX |
| Device | SNOMED |
| Observation | SNOMED |
| Visit | CMS Place of Service, ABMT, NUCC |
Concepts
The below image, taken from here, shows the IDs of all concepts in the CONCEPT table:
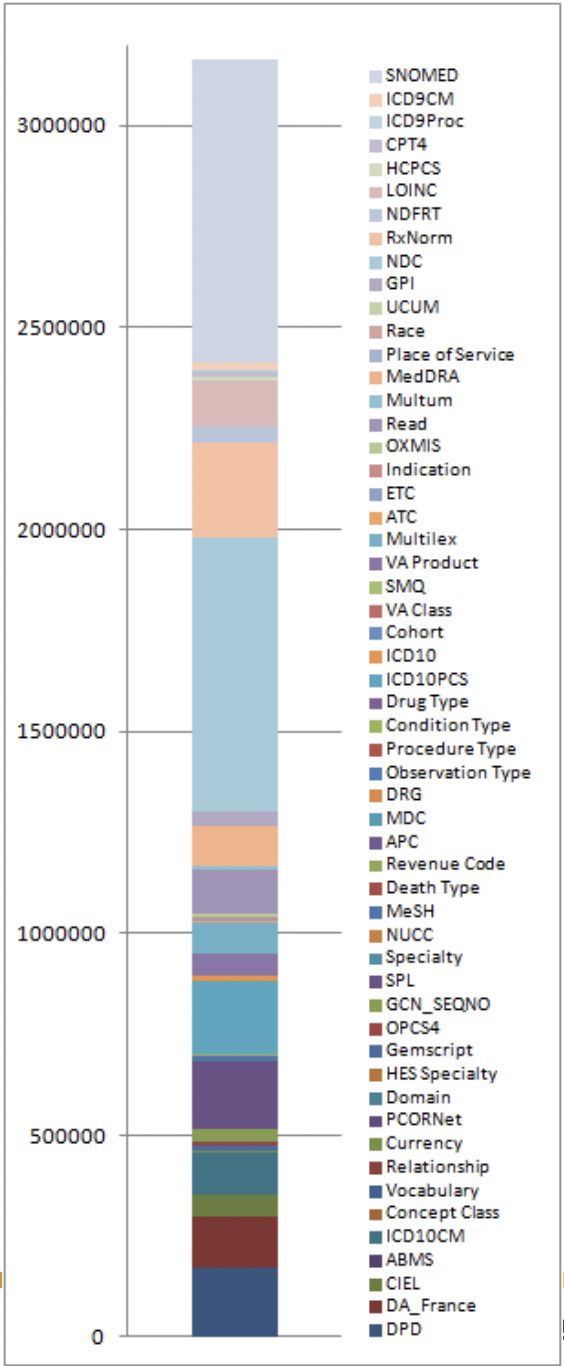
Note: Athena is a useful web GUI provided by OHDSI for searching through all OMOP concepts.
Relationships Between Concepts
Relationships between two concepts can exist across domains/vocabularies.
They are defined in the RELATIONSHIP table.
-
RELATIONSHIP_ID= primary key – unique alphanumeric string of <=20 chars, defines the type of relationship (e.g. “Maps to”, “Equivalent concepts” ) -
Linked to concepts as a foreign key on the
RELATIONSHIP_IDfield in the CONCEPT_RELATIONSHIP table
Mapping Source Concepts -> OMOP Standard Concepts
Here, we’ll walk through an example taken from the OHDSI training materials.
Goal: We’re given the ICD-9 concept 427.31 (atrial fibrilation). How do we get its corresponding standard OMOP concept?
Steps:
First, find the CONCEPT_ID in the CONCEPT table that corresponds to ICD-9 code 427.31
SELECT * FROM concept WHERE concept_code = '427.31'
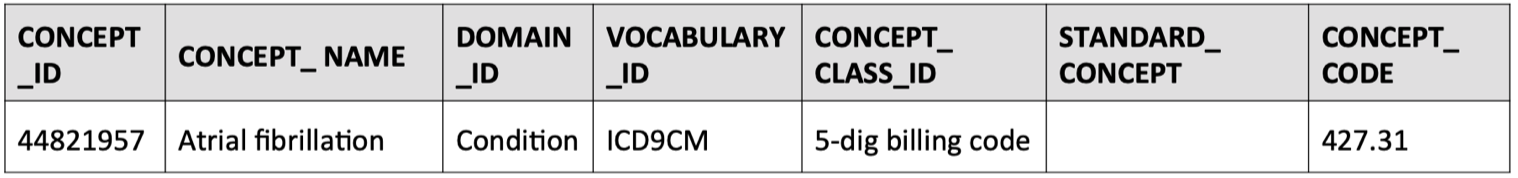
Here, we see that its correspondingCONCEPT_ID = 44821957
Second, map the CONCEPT_ID to its corresponding standard concept ID using the CONCEPT_RELATIONSHIP table.
Note that the RELATIONSHIP_ID = 'Maps to' maps a non-standard concept (labeled as concept_id_1 below) to its OMOP standard concept (labeled as concept_id_2)
SELECT * FROM concept_relationship WHERE concept_id_1 = 44821957 AND relationship_id = 'Maps to';
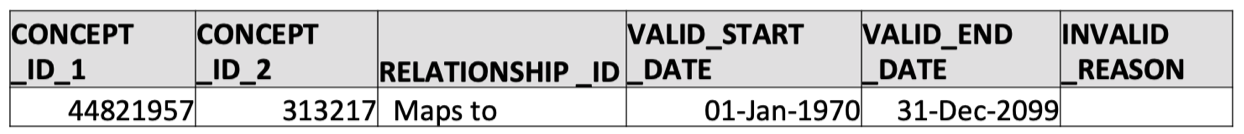
That’s it! We now know that the OMOP standard concept ID 313217 corresponds to ICD-9 code 427.31 (atrial fibrilation).
Generalizing:
Note that this process works for every concept and vocabulary.
We started with the ICD-9 code for atrial fibrilation (427.31), but we could also have used the Read code for atrial fibrilation (G573000), or the SNOMED code for atrial fibrilation (49436004).
The magic of OMOP is that all of these concepts will map to the same standard concept ID of 313217, which makes sense since they all refer to the same thing!
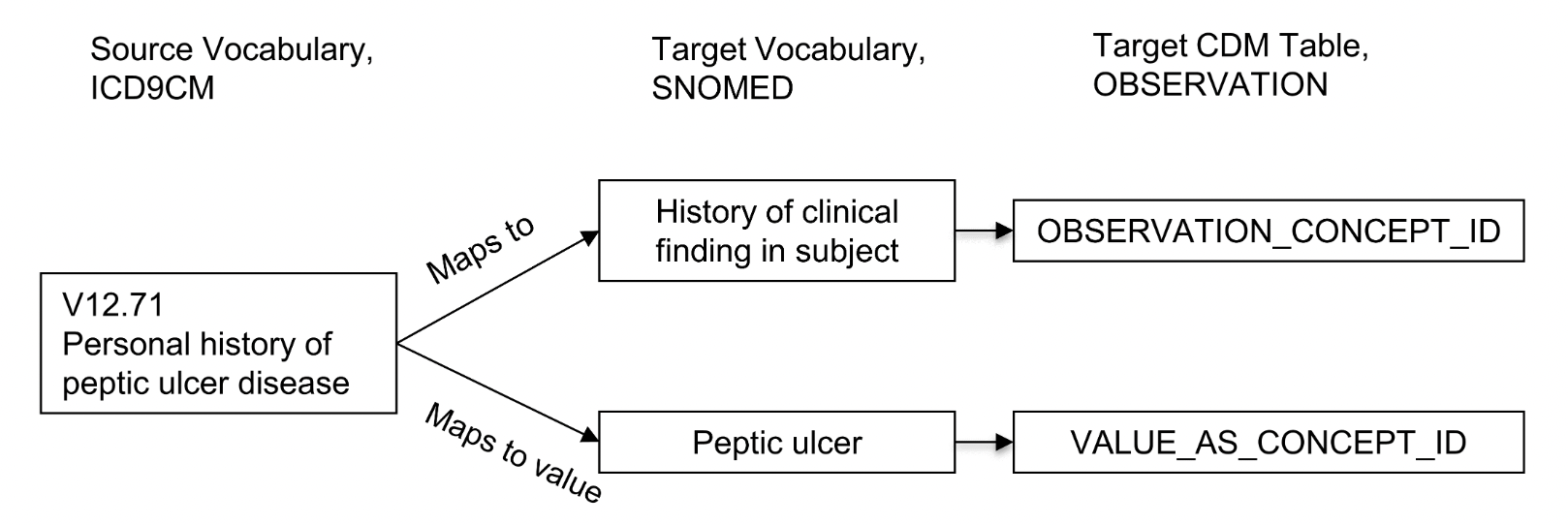
Mapping Lab Tests
One source concept can map to multiple concepts.
This is often the case for lab tests – the source concept will contain an attribute (e.g. the name of the test) and an associated value (e.g. the test result).
The source concept will have a “Maps to” relationship to the standard concept for the type of lab test, while the value will have a “Maps to Value” relationship to the standard concept for the test result.
The below image illustrates this mapping and is taken from here:
Field Naming Conventions
OMOP tables follow a standardized naming convention for some of their columns. This makes it simple to understand the relationships between tables.
The table below is taken from here and defines the meaning of columns in a given table named [Event]
| Notation | Description |
|---|---|
[Event]_ID |
Unique primary key. Usage: Foreign key for other Event tables. Example: PERSON_ID for patients, VISIT_OCCURRENCE_ID for Visits |
[Event]_CONCEPT_ID |
Foreign key to a Standard Concept in the CONCEPT table. Usage: Maps event to a normalized, unambiguous representation Example: CONDITION_CONCEPT_ID = 31967 references the SNOMED concept “Nausea” |
[Event]_SOURCE _CONCEPT_ID |
Foreign key to a Concept in the CONCEPT table. This is the equivalent of Source Value, and it may happen to be a Standard Concept, at which point it would be identical to the [Event]_CONCEPT_IDExample: CONDITION_SOURCE_CONCEPT_ID = 45431665 refernces the Read concept “Nausea” |
[Event]_TYPE_CONCEPT_ID |
Foreign key to a Concept in the CONCEPT table. Represents the capture mechanism that created this record. Example: DRUG_TYPE_CONCEPT_ID = "Pharmacy dispensing" if Drug was derived from a dispensing in the pharmacy, or "Prescription written" if derived from e-prescribing |
[Event]_SOURCE_VALUE |
Verbatim code or free text of this record in the source data. Example: CONDITION_SOURCE_VALUE = 78702 might correspond to ICD-9 code “787.02” written without a dot |
Read / Write Permissions
End-users can only write to the COHORT and COHORT_DEFINITION tables (from the Results category).
All of the other OMOP tables are read-only.
The only user that can write to these other tables is the ETL pipeline that the hospital uses to extract data out of their EHR and into OMOP.